Submission
1. Target genome of interest
• Select your target genome of interest.
• If there is no match, do select the most closest genome with yours.
• The selection of target genome is important for genome-wide off-target analysis.
2. Paste a sequence
• Sequence must be in FASTA format.
3. PAM sequence
• PAM (Protospacer Adjacent Motif)
• The choosing of PAM sequence is depending on the Cas9 used in experiment.
• For example, Streptococcus pyogenes Cas9 (SpCas9) recognizes NGG site (N stand for any nucleotide).
• Only NGG site is provided at the moment.
List of bacterial genomes
CRISPRBAC provides a wide range of bacterial species as a template for genome modification such as Salmonella, E. coli and Pseudomonas. Here is a total of 217 bacterial genomes that were kept in our database.
| # | Genus | Genome |
|---|---|---|
| 1 | Acinobacter | Acinobacter baumannii str. ACICU (NC_010611) |
| 2 | Aeromonas | Aeromonas hydrophila str. ML09-119 (NC_021290) |
| 3 | Aeromonas hydrophila subsp. hydrophila str. ATCC 7966 (NC_008570) | |
| 4 | Aeromonas salmonicida subsp. salmonicida str. A449 (NC_009348) | |
| 5 | Aeromonas veronii str. B565 (NC_015424) | |
| 6 | Anaplasma | Anaplasma centrale str. Israel (NC_013532) |
| 7 | Anaplasma marginale str. Florida (NC_012026) | |
| 8 | Anaplasma phagocytophilum str. HZ (NC_007797) | |
| 9 | Bacillus | Bacillus anthracis str. Ames (NC_003997) |
| 10 | Bacillus bacilliformis str. KC583 (NC_008783) | |
| 11 | Bacillus cereus str. ATCC 14579 (NC_004722) | |
| 12 | Bacillus cereus str. Q1 (NC_011969) | |
| 13 | Bacillus clausii str. KSM-K16 (NC_006582) | |
| 14 | Bacillus halodurans str. C-125 (NC_002570) | |
| 15 | Bacillus henselae str. Houston-1 (NC_005956) | |
| 16 | Bacillus licheniformis str. ATCC 14580 (NC_006270) | |
| 17 | Bacillus quintana str. Toulouse (NC_005955) | |
| 18 | Bacillus subtilis subsp. subtilis str. 168 (NC_000964) | |
| 19 | Bacillus thuringiensis serovar konkukian str. 97-27 (NC_005957) | |
| 20 | Bacillus tribocorum str. CIP 105476 (NC_010161) | |
| 21 | Bacillus weihenstephanensis str. KBAB4 (NC_010184) | |
| 22 | Bordetella | Bordetella avium str. 197N (NC_010645) |
| 23 | Bordetella bronchiseptica str. RB50 (NC_002927) | |
| 24 | Bordetella parapertussis str. 12822 (NC_002928) | |
| 25 | Bordetella pertussis str. Tohama I (NC_002929) | |
| 26 | Brucella | Brucella abortus bv. 1 str. 9-941 chromosome I (NC_006932) |
| 27 | Brucella abortus bv. 1 str. 9-941 chromosome II (NC_006933) | |
| 28 | Brucella canis str. ATCC 23365 chromosome I (NC_010103) | |
| 29 | Brucella canis str. ATCC 23365 chromosome II (NC_010104) | |
| 30 | Brucella melitensis biovar Abortus str. 2308 chromosome I (NC_007618) | |
| 31 | Brucella melitensis biovar Abortus str. 2308 chromosome II (NC_007624) | |
| 32 | Brucella melitensis bv. 1 str. 16M chromosome I (NC_003317) | |
| 33 | Brucella melitensis bv. 1 str. 16M chromosome II (NC_003318) | |
| 34 | Brucella melitensis str. ATCC 23457 chromosome I (NC_012441) | |
| 35 | Brucella melitensis str. ATCC 23457 chromosome II (NC_012442) | |
| 36 | Brucella ovis str. ATCC 25840 chromosome I (NC_009505) | |
| 37 | Brucella ovis str. ATCC 25840 chromosome II (NC_009504) | |
| 38 | Brucella suis str. 1330 chromosome I (NC_004310) | |
| 39 | Brucella suis str. 1330 chromosome II (NC_004311) | |
| 40 | Burkholderia | Burkholderia cenocepacia str. J2315 chromosome I (NC_011000) |
| 41 | Burkholderia cenocepacia str. J2315 chromosome II (NC_011001) | |
| 42 | Burkholderia cenocepacia str. J2315 chromosome III (NC_011002) | |
| 43 | Burkholderia mallei str. ATCC 23344 chromosome I (NC_006348) | |
| 44 | Burkholderia mallei str. ATCC 23344 chromosome II (NC_006349) | |
| 45 | Burkholderia pseudomallei str. K96243 chromosome I (NC_006350) | |
| 46 | Burkholderia pseudomallei str. K96243 chromosome II (NC_006351) | |
| 47 | Burkholderia thailandensis str. E264 chromosome I (NC_007651) | |
| 48 | Burkholderia thailandensis str. E264 chromosome II (NC_007650) | |
| 49 | Campylobacter | Campylobacter fetus subsp. fetus str. 82-40 (NC_008599) |
| 50 | Campylobacter jejuni str. RM1221 (NC_003912) | |
| 51 | Campylobacter jejuni subsp. doylei str. 269.97 (NC_009707) | |
| 52 | Campylobacter jejuni subsp. jejuni str. 81116 (NC_009839) | |
| 53 | Chlamydia | Chlamydia abortus str. S26/3 (NC_004552) |
| 54 | Chlamydia caviae str. GPIC (NC_003361) | |
| 55 | Chlamydia felis str. Fe/C-56 (NC_007899) | |
| 56 | Chlamydia muridarum str. Nigg (NC_002620) | |
| 57 | Chlamydia pneumoniae str. J138 (NC_002491) | |
| 58 | Chlamydia trachomatis str. 434/Bu (NC_010287) | |
| 59 | Chlamydia trachomatis str. A/HAR-13 (NC_007429) | |
| 60 | Chlamydia trachomatis str. D/UW-3/CX (NC_000117) | |
| 61 | Chlamydia trachomatis str. L2b/UCH-1/proctitis (NC_010280) | |
| 62 | Clostridium | Clostridium acetobutylicum str. ATCC 824 (NC_003030) |
| 63 | Clostridium beijerinckii str. NCIMB 8052 (NC_009617) | |
| 64 | Clostridium botulinum A str. ATCC 3502 (NC_009495) | |
| 65 | Clostridium botulinum A2 str. Kyoto (NC_012563) | |
| 66 | Clostridium botulinum A3 str. Loch Maree (NC_010520) | |
| 67 | Clostridium botulinum B str. Eklund 17B (NC_010674) | |
| 68 | Clostridium botulinum B1 str. Okra (NC_010516) | |
| 69 | Clostridium botulinum E3 str. Alaska E43 (NC_010723) | |
| 70 | Clostridium botulinum F str. Langeland (NC_009699) | |
| 71 | Clostridium difficile str. 630 (NC_009089) | |
| 72 | Clostridium novyi str. NT (NC_008593) | |
| 73 | Clostridium perfringens str. ATCC 13124 (NC_008261) | |
| 74 | Clostridium tetani str. E88 (NC_004557) | |
| 75 | Clostridium thermocellum str. ATCC 27405 (NC_009012) | |
| 76 | Corynebacterium | Corynebacterium diphtheriae str. NCTC 13129 (NC_002935) |
| 77 | Corynebacterium efficiens str. YS-314 (NC_004369) | |
| 78 | Corynebacterium glutamicum str. ATCC 13032 (NC_003450) | |
| 79 | Corynebacterium jeikeium str. K411 (NC_007164) | |
| 80 | Corynebacterium pseudotuberculosis str. FRC41 (NC_014329) | |
| 81 | Coxiella | Coxiella burnetii str. RSA 493 (NC_002971) |
| 82 | Enterococcus | Enterococcus faecalis str. D32 (NC_018221) |
| 83 | Enterococcus faecalis str. V583 (NC_004668) | |
| 84 | Enterococcus faecium str. DO (NC_017960) | |
| 85 | Escherichia | Escherichia |
| 86 | Escherichia coli O104:H4 str. 2009EL-2050 (NC_018650) | |
| 87 | Escherichia coli O157:H7 str. Sakai (NC_002695) | |
| 88 | Escherichia coli O25b:H4-ST131 strain:EC958 (NZ_HG941718) | |
| 89 | Escherichia coli O26:H11 str. 11368 (NC_013361) | |
| 90 | Escherichia coli O44:H18 str. 042 (NC_017626) | |
| 91 | Escherichia coli O55:H7 str. RM12579 (NC_017656) | |
| 92 | Escherichia coli O7:K1 str. CE10 (NC_017646) | |
| 93 | Escherichia coli O83:H1 str. NRG 857C (NC_017634) | |
| 94 | Escherichia coli str. 536 (NC_008253) | |
| 95 | Escherichia coli str. APEC O1 (NC_008563) | |
| 96 | Escherichia coli str. K-12 substr. MG1655 (NC_000913) | |
| 97 | Escherichia coli str. SMS-3-5 (NC_011745) | |
| 98 | Escherichia coli str. UMNK88 (NC_017641) | |
| 99 | Francisella noatunensis subsp. orientalis str. Toba 04 (NC_017909) | |
| 100 | Francisella | Francisella philomiragia subsp. philomiragia str. ATCC 25017 (NC_010336) |
| 101 | Francisella tularensis subsp. holarctica str. FSC200 (NC_019551) | |
| 102 | Francisella tularensis subsp. mediasiatica str. FSC147 (NC_010677) | |
| 103 | Francisella tularensis subsp. tularensis str. SCHU S4 (NC_006570) | |
| 104 | Haemophilus | Haemophilus ducreyi 35000HP (AE017143.1) |
| 105 | Haemophilus influenzae PittEE (CP000671.1) | |
| 106 | Haemophilus influenzae Rd KW20 (L42023.1) | |
| 107 | Haemophilus somnus 129PT (CP000436.1) | |
| 108 | Helicobacter | Helicobacter acinonychis str. Sheeba (AM260522.1) |
| 109 | Helicobacter hepaticus ATCC 51449 (AE017125.1) | |
| 110 | Helicobacter mustelae 12198 (FN555004.1) | |
| 111 | Helicobacter pylori 26695 (AE000511.1) | |
| 112 | Klebsiella | Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 (AP006725.1) |
| 113 | Klebsiella variicola strain 342 (CP000964.1) | |
| 114 | Legionella | Legionella longbeachae NSW150 (FN650140.1) |
| 115 | Legionella pneumophila subsp. pneumophila str. Philadelphia 1 (AE017354.1) | |
| 116 | Listeria | Listeria innocua Clip11262 (AL592022.1) |
| 117 | Listeria ivanovii subsp. ivanovii PAM 55 (NC_016011.1) | |
| 118 | Listeria monocytogenes EGD-e (AL591824.1) | |
| 119 | Listeria monocytogenes serotype 7 str. SLCC2482 (FR720325.1) | |
| 120 | Listeria monocytogenes str. 4b F2365 (AE017262.2) | |
| 121 | Listeria monocytogenes strain SLCC2372, serotype 1/2c (FR733648.1) | |
| 122 | Listeria monocytogenes strain SLCC2755, serotype 1/2b (FR733646.1) | |
| 123 | Listeria seeligeri serovar 1/2b str. SLCC3954 (FN557490.1) | |
| 124 | Listeria welshimeri serovar 6b str. SLCC5334 (AM263198.1) | |
| 125 | Mycobacterium | Mycobacterium abscessus subsp. massiliense str. GO 06 (CP003699.2) |
| 126 | Mycobacterium africanum GM041182 (FR878060.1) | |
| 127 | Mycobacterium avium subsp. paratuberculosis str. k10 (AE016958.1) | |
| 128 | Mycobacterium bovis BCG str. Mexico (CP002095.1) | |
| 129 | Mycobacterium canettii CIPT 140060008 (FO203507.1) | |
| 130 | Mycobacterium gilvum Spyr1 (CP002385.1) | |
| 131 | Mycobacterium indicus pranii MTCC 9506 (CP002275.1) | |
| 132 | Mycobacterium tuberculosis H37Rv (AL123456.3) | |
| 133 | Mycobacterium ulcerans Agy99 (CP000325.1) | |
| 134 | Mycoplasma | Mycoplasma agalactiae PG2 (CU179680.1) |
| 135 | Mycoplasma arthritidis 158L3-1 (CP001047.1) | |
| 136 | Mycoplasma capricolum subsp. capricolum ATCC 27343 (CP000123.1) | |
| 137 | Mycoplasma conjunctivae HRC/581T (NC_012806.1) | |
| 138 | Mycoplasma fermentans JER (CP001995.1) | |
| 139 | Mycoplasma gallisepticum str. R(low) (AE015450.2) | |
| 140 | Mycoplasma genitalium G37 (L43967.2) | |
| 141 | Mycoplasma hominis ATCC 23114 (FP236530.1) | |
| 142 | Mycoplasma hyopneumoniae 232 (AE017332.1) | |
| 143 | Mycoplasma mobile 163K (AE017308.1) | |
| 144 | Mycoplasma mycoides subsp. mycoides SC str. PG1 (BX293980.2) | |
| 145 | Mycoplasma penetrans HF-2 DNA (BA000026.2) | |
| 146 | Mycoplasma pneumoniae M129 (U00089.2) | |
| 147 | Mycoplasma pulmonis UAB CTIP (AL445566.1) | |
| 148 | Mycoplasma synoviae 53 (AE017245.1) | |
| 149 | Neisseria | Neisseria gonorrhoeae NCCP11945 (CP001050.1) |
| 150 | Neisseria lactamica 020-06 (FN995097.1) | |
| 151 | Neisseria meningitidis MC58 (AE002098.2) | |
| 152 | Pseudomonas | Pseudomonas aeruginosa PAO1 (AE004091.2) |
| 153 | Pseudomonas entomophila str. L48 (CT573326.1) | |
| 154 | Pseudomonas fluorescens SBW25 (AM181176.4) | |
| 155 | Pseudomonas mendocina ymp (CP000680.1) | |
| 156 | Pseudomonas putida KT2440 (AE015451.2) | |
| 157 | Pseudomonas savastanoi pv. phaseolicola 1448A (NC_005773.3) | |
| 158 | Pseudomonas stutzeri A1501 (CP000304.1) | |
| 159 | Rickettsia | Rickettsia africae ESF-5 (CP001612.1) |
| 160 | Rickettsia akari str. Hartford (CP000847.1) | |
| 161 | Rickettsia bellii OSU 85-389 (CP000849.1) | |
| 162 | Rickettsia canadensis str. McKiel (CP000409.1) | |
| 163 | Rickettsia conorii str. Malish 7 (AE006914.1) | |
| 164 | Rickettsia massiliae MTU5 (CP000683.1) | |
| 165 | Rickettsia peacockii str. Rustic (CP001227.1) | |
| 166 | Rickettsia prowazekii str. Madrid E (AJ235269.1) | |
| 167 | Rickettsia rickettsii str. Iowa (CP000766.3) | |
| 168 | Rickettsia typhi str. Wilmington (AE017197.1) | |
| 169 | Salmonella | Salmonella enterica subsp. arizonae serovar 62:z4,z23:-- (NC_010067.1) |
| 169 | Salmonella enterica subsp. enterica serovar Agona str. SL483 (CP001138.1) | |
| 170 | Salmonella enterica subsp. enterica serovar Choleraesuis str. SC-B67 (AE017220.1) | |
| 171 | Salmonella enterica subsp. enterica serovar Dublin str. CT_02021853 (CP001144.1) | |
| 172 | Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 (AM933172.1) | |
| 173 | Salmonella enterica subsp. enterica serovar Gallinarum str. 287/91 (AM933173.1) | |
| 174 | Salmonella enterica subsp. enterica serovar Heidelberg str. SL476 (CP001120.1) | |
| 175 | Salmonella enterica subsp. enterica serovar Newport str. SL254 (CP001113.1) | |
| 176 | Salmonella enterica subsp. enterica serovar Paratyphi A str. AKU_12601 (FM200053.1) | |
| 177 | Salmonella enterica subsp. enterica serovar Paratyphi A str. ATCC 9150 (CP000026.1) | |
| 178 | Salmonella enterica subsp. enterica serovar Paratyphi B str. SPB7 (CP000886.1) | |
| 179 | Salmonella enterica subsp. enterica serovar Paratyphi C strain RKS4594 (NC_012125.1) | |
| 180 | Salmonella enterica subsp. enterica serovar Schwarzengrund str. CVM19633 (CP001127.1) | |
| 181 | Salmonella enterica subsp. enterica serovar Typhi str. CT18 (AL513382.1) | |
| 182 | Salmonella enterica subsp. enterica serovar Typhi Ty2 (AE014613.1) | |
| 183 | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (AE006468.2) | |
| 184 | Shigella | Shigella boydii Sb227 (CP000036.1) |
| 185 | Shigella dysenteriae Sd197 (CP000034.1) | |
| 186 | Shigella flexneri 2a str. 2457T (AE014073.1) | |
| 187 | Shigella flexneri 5 str. 8401 (CP000266.1) | |
| 188 | Shigella sonnei Ss046 (CP000038.1) | |
| 189 | Staphylococcus | Staphylococcus aureus RF122 (AJ938182.1) |
| 190 | Staphylococcus aureus subsp. aureus Mu3 (AP009324.1) | |
| 191 | Staphylococcus aureus subsp. aureus Mu50 (BA000017.4) | |
| 192 | Staphylococcus epidermidis RP62A (CP000029.1) | |
| 193 | Staphylococcus haemolyticus JCSC1435 (AP006716.1) | |
| 194 | Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 (AP008934.1) | |
| 195 | Streptococcus | Streptococcus agalactiae A909 (CP000114.1) |
| 196 | Streptococcus dysgalactiae subsp. equisimilis GGS_124 (AP010935.1) | |
| 197 | Streptococcus equi subsp. equi 4047 (FM204883.1) | |
| 198 | Streptococcus equi subsp. zooepidemicus MGCS10565 (CP001129.1) | |
| 199 | Streptococcus gordonii str. Challis substr. CH1 (CP000725.1) | |
| 200 | Streptococcus mutans NN2025 (AP010655.1) | |
| 201 | Streptococcus pneumoniae R6 (AE007317.1) | |
| 202 | Streptococcus pyogenes MGAS5005 (CP000017.2) | |
| 203 | Streptococcus sanguinis SK36 (CP000387.1) | |
| 204 | Streptococcus suis 05ZYH33 (CP000407.1) | |
| 205 | Streptococcus thermophilus LMD-9 (CP000419.1) | |
| 206 | Vibrio | Vibrio cholerae O1 biovar eltor str. N16961 chromosome I (AE003852.1) |
| 207 | Vibrio cholerae O1 biovar eltor str. N16961 chromosome II (AE003853.1) | |
| 208 | Vibrio fischeri ES114 chromosome I (CP000020.2) | |
| 209 | Vibrio fischeri ES114 chromosome II (CP000021.2) | |
| 210 | Vibrio parahaemolyticus RIMD 2210633 (BA000031.2) | |
| 211 | Vibrio parahaemolyticus RIMD 2210633 (BA000032.2) | |
| 212 | Vibrio vulnificus CMCP6 chromosome I (AE016795.3) | |
| 213 | Vibrio vulnificus CMCP6 chromosome II (AE016796.2) | |
| 214 | Yersinia | Yersinia enterocolitica subsp. enterocolitica 8081 (AM286415.1) |
| 215 | Yersinia enterocolitica subsp. palearctica 105.5R(r) (NC_015224.1) | |
| 216 | Yersinia pestis biovar Microtus str. 91001 (AE017042.1) | |
| 217 | Yersinia pseudotuberculosis IP 31758 (CP000720.1) |
On-target activity scoring
On-target is the ability of gRNA to successfully find and bind Cas9 to the target sequence (Mendoza & Trinh, 2018).
On-target activity is scored based on physical and sequence features of gRNA such as GC content,
minimum free energy (MFE), melting temperature (Tm), position-specific nucleotide and non-specific
position nucleotide. Of 479 features, we used Random Forest to select important features by comparing 5-fold cross-validation
results of top 50, 100, 150, 200, 250, 300, 350, 400 and 479 features. The features were then ranked based
on the mean decrease impurity (Gini importance). We found that the top 150 features have higher accuracy
of predicted model (Accuracy = 0.7316). Then, we used Logistic Regression coefficient to obtain model weight
for each feature (Accuracy = 0.7353). This model weight will be further used to compute on-target scoring.
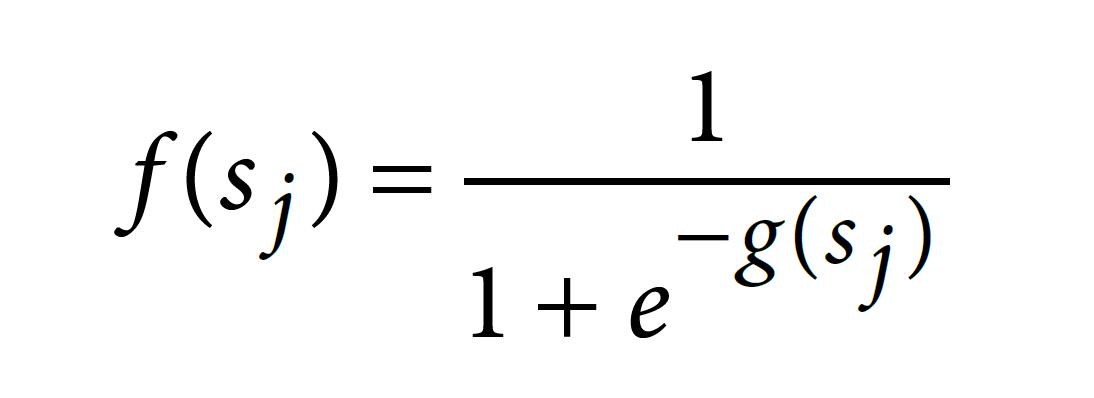
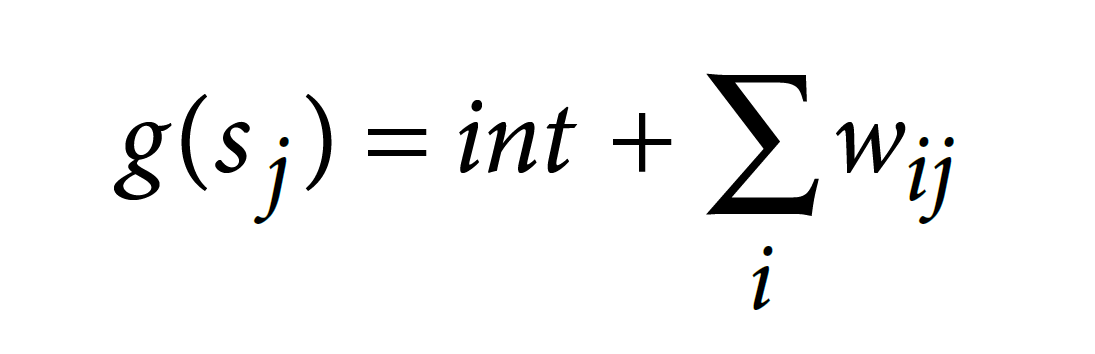
• int (intercept) = 0.0412385
• wij = weight of feature i
The score is ranged from 0 to 1. Higher score indicates higher predicted on-target activity.
Off-target scoring
Off-target is the probabilty of gRNA sequence bind to a non-matching sequence in the genome (Mendoza & Trinh, 2018).
Our off-target score use CFD (Cutting Frequency Determination) score generated by Doench et. al., (2016) to
predict off-target activity of gRNAs. For more details, please refer to their publication.
The score is ranged from 0 to 1. Higher score indicates higher off-target activity
(Note: Best gRNA would have lower off-target score).
Interpreting results
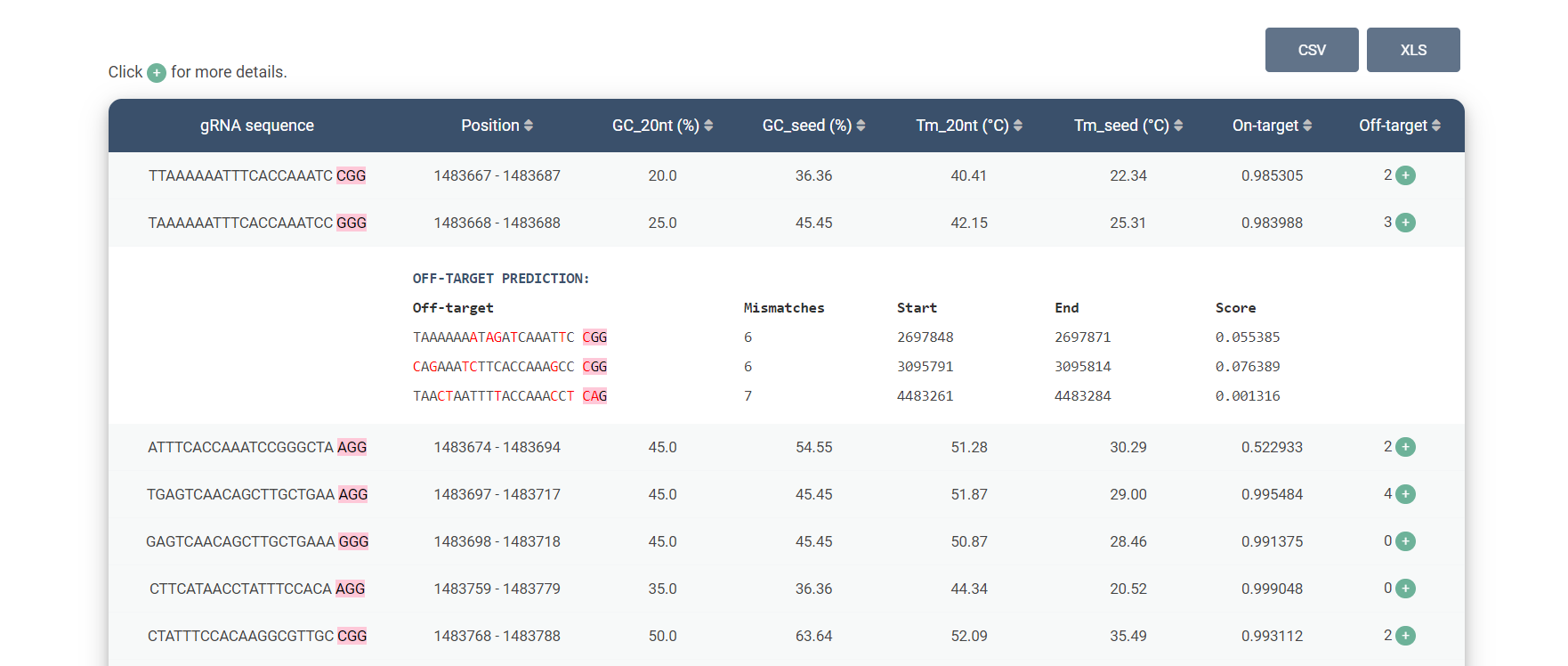
Overview results:
| Items | Description |
|---|---|
| gRNA sequence | 20-nucleotide sequence with PAM (highlighted). |
| Position | Position of gRNA in selected target genome. |
| GC_20nt | GC content of entire gRNA sequence. |
| GC_seed | GC content of seed region (10th to 20th bases). |
| Tm_20nt | Melting temperature of entire gRNA sequence. |
| Tm_seed | Melting temperature of seed region. |
| On-target |
|
| Off-target |
|
Off-target prediction table:
| Items | Description |
|---|---|
| Off-target | 20-nucleotide off-target sequence with PAM (highlighted). |
| Mismatches | Number of mismatches. |
| Start | Start position of off-target in target genome. |
| End | End position of off-target in target genome. |
| CFD score |
|
References
Mendoza, B. J., & Trinh, C. T. (2018). Enhanced guide-RNA design and targeting analysis for precise CRISPR genome
editing of single and consortia of industrially relevant and non-model organisms. Bioinformatics, 34(1), 16-23.
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., ... & Virgin, H. W. (2016).
Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nature biotechnology, 34(2), 184.